scOrange Blog
-
Categories:
- clustering
- t-sne
- marker-genes
- tsne
- annotator
- data-mining
- embedding
- embryo
- genomics
- markers
Author: Iva Černoša, Jun 6, 2019
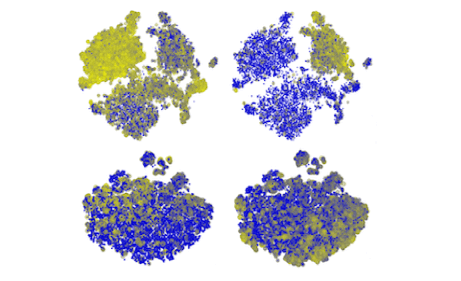
Identification of Cell Populations in Healthy Bone Marrow
We identify cell populations in a dataset of healthy human bone marrow cells from Galen et al. (Cell, 2019) and take a look at the putative cell differentiation trajectories.
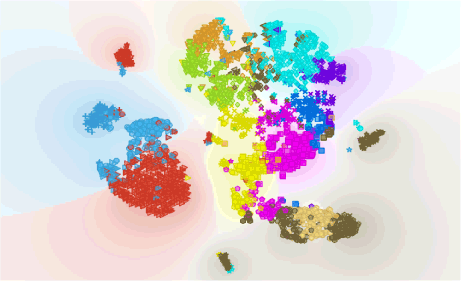
Author: Iva Černoša, May 10, 2019
Pinpointing Lineage Differentiation in Human Preimplantation Embryotic Cells
Combine clustering and the time component to pinpoint the time of trophectoderm (TE) and inner cell mass (ICM) differentiation in human preimplantation embryotic cells and recreate a part of study by Petropoulos et al. (Cell, 2016)
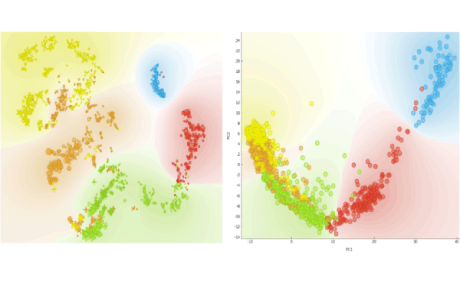
Author: Iva Černoša, May 6, 2019
X And Y Chromosome Expression in Human Preimplantation Embryos
Using the single cell data from Petropoulos et al. (Cell, 2016), we determine the karyotype (XY, XX, X0) of the cells and inspect the time of activation of Y chromosome in a preimplantation embryo.
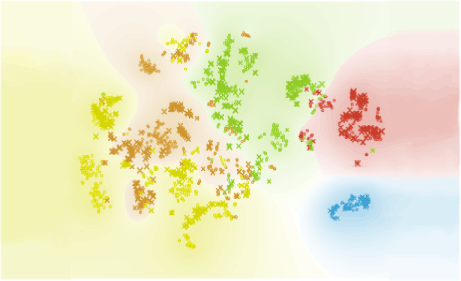
Author: Iva Černoša, Apr 23, 2019
Subsets of Exhausted CD8+ T Cells
We reproduce a part of the study by Miller et al. (Nature Immunology, 2019) through identifying subsets of exhausted CD8+ T cells during chronic viral infections and corresponding groups in T cells isolated from tumours